
 |
Manual |
In this module you will find a tool for selecting a best matching model among a series of models, by comparing their pre-calculated hydrodynamic parameters with user-provided experimental values.
 |
|
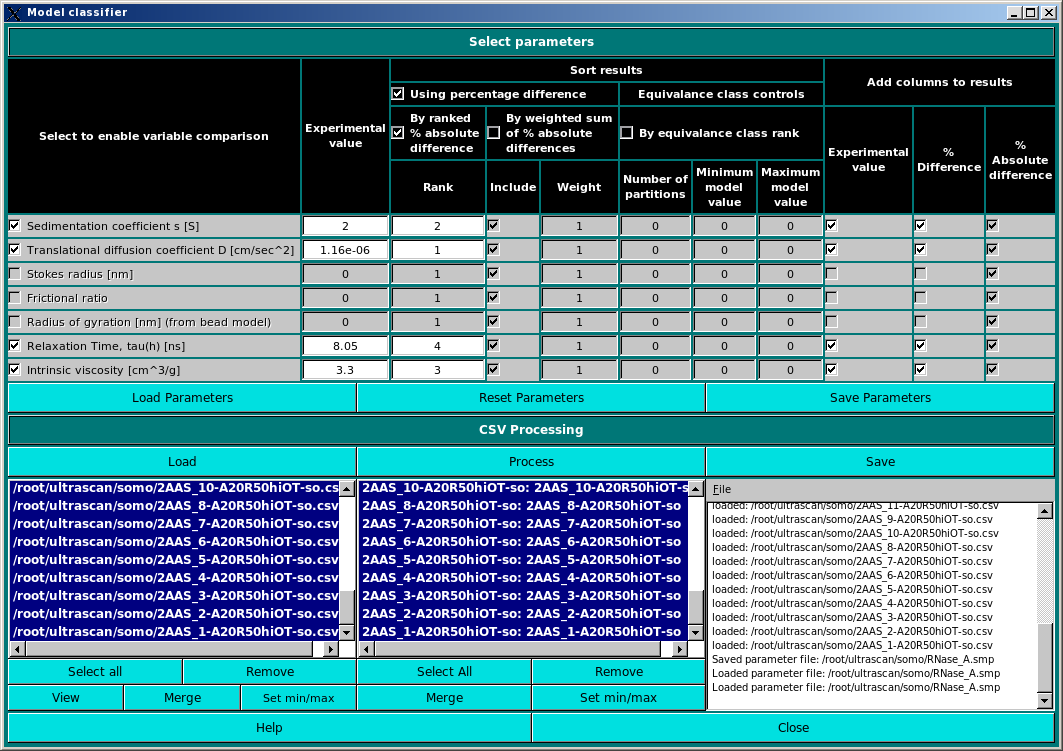
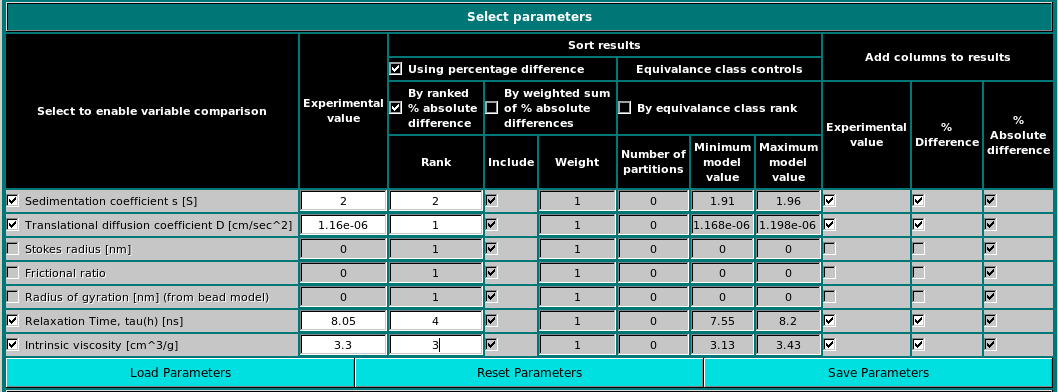
In the first panel, the experimental parameters to be used are entered, and the methods used to screen the computed results against them are set.
The currently available parameters are listed in the Select to enable variable comparison column, and include:
Enter then the experimental values in the relative fields in the second column, Experimental value. Be careful to use the appropriate units, as indicated after each parameter in the first column. The results can be sorted using several alternative criteria, listed under the Sort results group:
The Model classifier will write the results in a file. Under the Add columns to results header you will find three columns containing checkboxes, Experimental value, % Difference and % Absolute difference. Selecting one or more checkboxes will add the corresponding value(s) to the output file.
|
 |
|
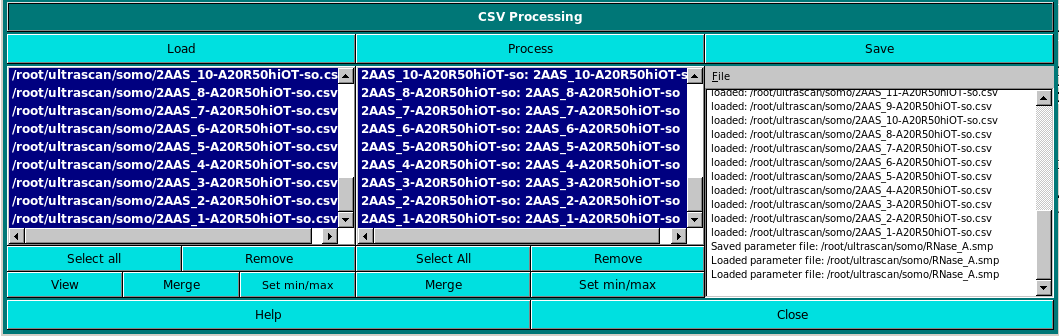
In the second panel, the calculated parameters are uploaded. They should be in *.csv files, most easily generated using the Save parameters to a file checkbox (and the Select Parameters to be Saved module) in the Hydrodynamic calculations section of the main US-SOMO window. Only the parameters present in the *.csv files, identified through their headers, will be then available in the Select to enable variable comparison column.
Pressing the Load button will open the filesystem dialog and allow importing the required *.csv files into the left-side window. Files can then be selected by clicking on each filename, which will transfer them to the right-side window, or by pressing the Select all button.
The files listed in the right-side window can be then selected for processing by individually clicking on them, or by pressing the Select all button.
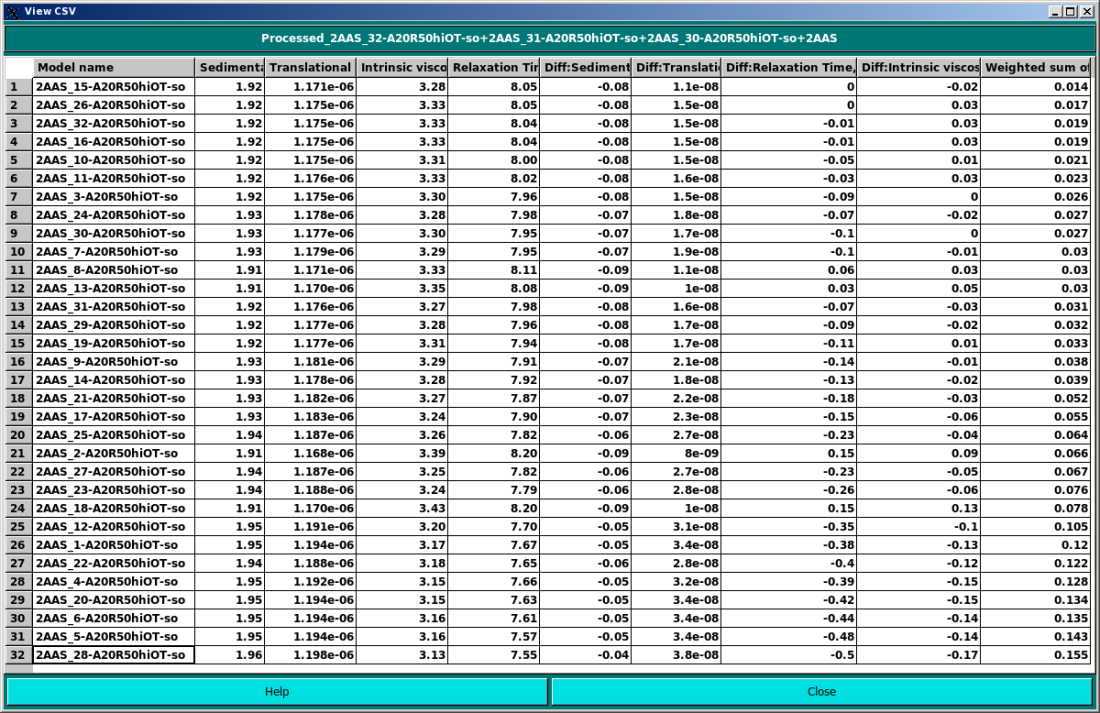
Once files has been selected, the Model Classifier can be launched by pressing the Process button, and the progress window at the far right will be updated. At the end, pressing View will open a window with all the selected columns:
|
 |
|
In the example above, the classifier used the weighted sum of % absolute differences to rank the models. By clicking on each column, the results can be sorted according to that column's values. By pressing the Save button, a filesystem dialog will open allowing to save the results in a new *.csv file, which can then be opened with a standard spreadsheet. A typical results file after ranking by % absolute difference can be seen below: |
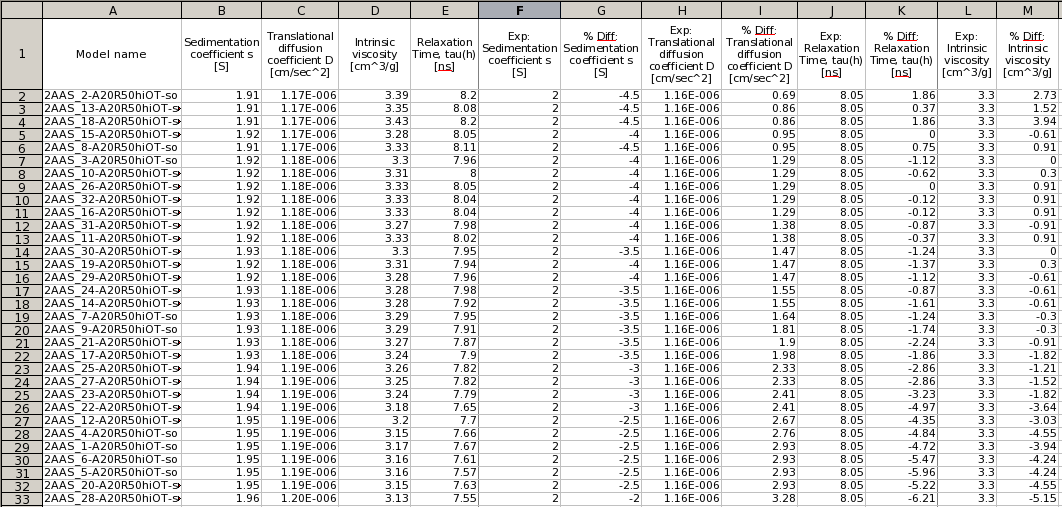 |
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on March 16, 2012.